
|
||||||||
| Genomic Services | ||||||||
|
||||||||
| Protein Services | ||||||||
|
||||||||
Located in rooms
B065 and B017 |
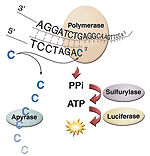
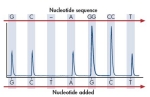
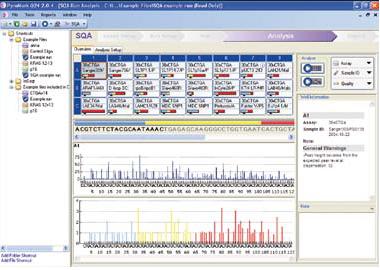
| Pyrosequencing |
• Sample Submission |
** Please check for the appropriate sample submission requirements for each service type before placing your orders
Full Service - PCR and Pyrosequencing Run: (back to top)
|
Pyrosequencing Run Only: (back to top)
|
Self Service: (back to top)
|
Home | FAQs | | Prices | Contact Us | Publications | Feedback
Beckman Center | Stanford Medical Center | Stanford University
© 2006 Stanford PAN Facility. All rights reserved