- Where is the PAN Facility located/contact info?
- What is the turnaround time?
- How much does it cost?
- What primers does the PAN Facility provide?
- What concentration should the DNA be?
- What concentration should the primers be?
- How do I get my results?
- Why do I need three primers?
- Assay Design tips
- PCR using PyroMark PCR Kit
1. Where is the PAN Facility located/contact info? (top)
We are located in the basement of the Beckman Center, B017. We also have a drop-off area in cold room B024.
Phone: (650) 723-3189
Email: panpyro-qpcr@stanford.edu
Shipping Address:
Stanford PAN Facility
Beckman Center, B017
279 Campus Drive, West
Stanford, Ca 94305
2. What is the turnaroud time? (top)
Turnaround time is project dependent. On average assay design take a week. Optimizing PCR can take a week. Once optimal PCR conditions have been established, pyrosequencing samples are processed in ~two days.
3. How much does it cost? (top)
Please check our current price list.
4. What primers does the PAN Facility provide? (top)
Unfortunately, the PAN Facility does not provide any primers for Pyrosequencing. We are working on establishing a universal PCR primer set which we hope will be available in the near future.
5. What concentration should the DNA be? (top)
In general, it is best to follow the your PCR kit's manufacturer's guidelines. Typically, we have seen good results with 10ng - 20ng of total DNA per PCR rxn.
6. What concentration should the primers be? (top)
Both PCR and Pyro-Sequencing primers should be at an initial concentration of 5uM. Consider supplying 5ul of each primer for each rxn.
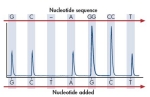
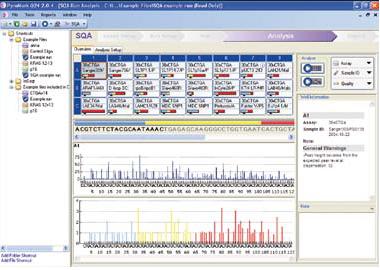
7. How do I get my results? (top)
Results are emailed to you, and can be viewed using Qiagen's PyroMark software. You can obtain a free copy of the software by contacting panpyro-qpcr@stanford.edu at 650-723-3189.
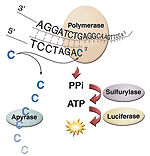
8. Why do I need three primers? (top)
You need at least two primers for PCR. One PCR primer MUST BE biotinylated on the 5' end. The biotinlyated PCR primer can be either the forward or reverse PCR primer, that's up to you. The biotinlyated PCR strand is captured/purified prior to pyrosequencing. A pyro-sequencing primer (the third primer) is added to the purified biotinlylated PCR strand and pyrosequencing commenses. The pryo-sequencing primer MUST BE in the opposite direction of the biotinlyated primer. You can use the non-biotinlyated PCR primer as your pyro-sequencing primer, but please be aware that this strategy is not as efficent as using a nested pyro-sequencing primer.
9. Assay Design Tips (top)
- Primers for PCR:
* The optimal PCR amplicon lengh is between 80 and 200bp, although 500bp might work well for Pyrosequencing assays on genomic DNA.
* TARGET REGION should be less than 100 bases.
* Avoid long stretches of homopolyers, especially next to variable regions (SNP/CpG sites).
* Avoid strong hairpins and primer dimers.
* SNP PCR primers should be 18-25bp, with annealing temp of 60-70°C.
* CpG PCR primers should be 22-30bp.
* One primer must be biotinylated and in the opposite direction of the Pyrosequencing Primer
* All primers should be purifired
** Specific SNP and CpG assay design guidelines can be found within the software's help menu.
- Primer for Pyrosequencing:
* Pyrosequencing primer should be 15-20 bases long, with annealing temp of 45-55°C.
* Pyrosequencing primer should be located 1-5 bases away from first SNP or CpG site.
- Ready-Made Assays for pyrosequencing are available from Qiagen.
* The assays consist of PCR primers and pyrosequencing primers designed for use with PyroMark systems. ~60,000 PyroMark CpG Assays for human, mouse, and rat gene sequences are available in convenient tube or 96-well formats. Custom-designed assays can also be ordered through Qiagen.
- Custom Assays should be designed using Qiagen's PyroMark Assay Design software.
* The software will generate PCR and pyrosequencing primers optimized for use with the PyroMark Q24. The software is available for FREE to our customers. To obtain a copy please contact us. We can provide assistance with training and design of pyrosequencing assays.
|
10. PCR using PyroMark PCR Kit (top)
- Follow instructions in the PyroMark PCR Handbook. Final volume for the PCR product is 25ul.
- Template DNA concentration: <500 ng/rxn or ~10-20ng/ul bisulfite converted DNA. We recommend using 1-10ng human genomic DNA.
- Primer concentration: 5uM
|
|