** Below are examples of the most common errors found in Bioanalyzer traces. This guide will help you understand your data better. If you have any questions or concerns about your data, please contact us at panbioanalyzer@stanford.edu.
** For further analysis of your samples, you can use the Agilent 2100 Expert Software to open the raw (.xad) data file.
RNA Assays - (back to top)
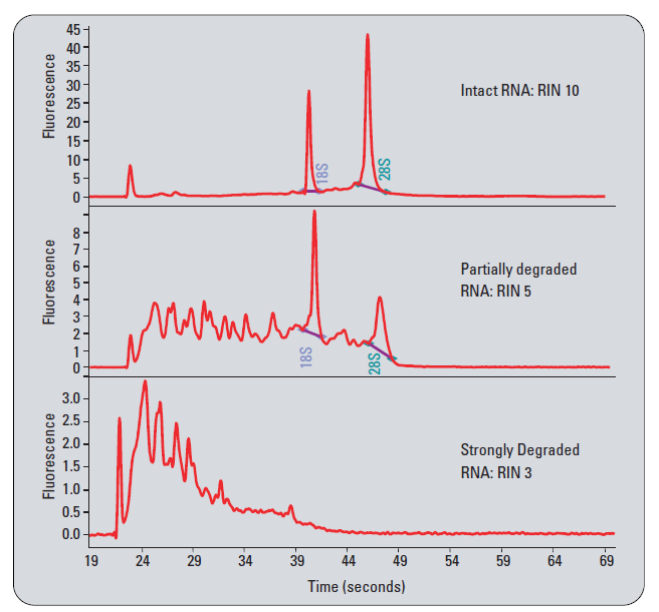
| • Total RNA Integrity – |
|
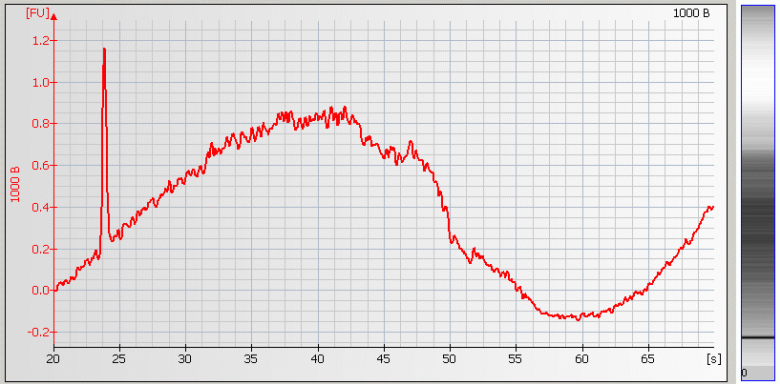
• Spikes – Microparticles and/or air bubbles in the system |
|
|
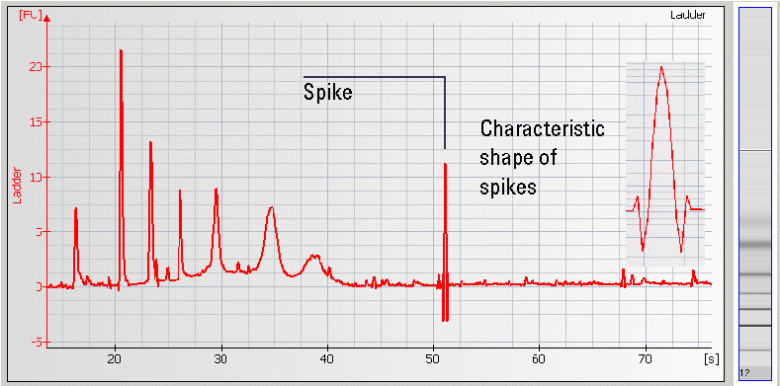
|
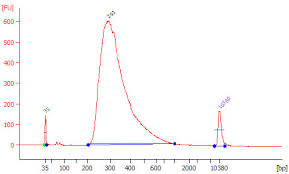
| • Broad Peaks – Sample contaminated with genomic DNA |
|
• Missing Peaks – Sample salt concentration is too high
|
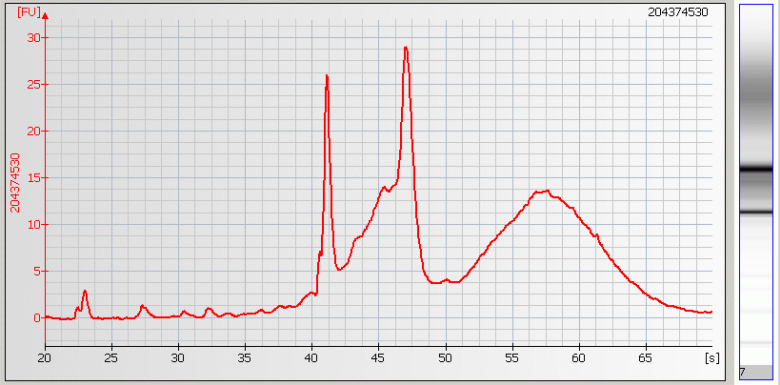
|
|
|
| • Missing RNA Fragments – Sample salt concentration is too high |
|
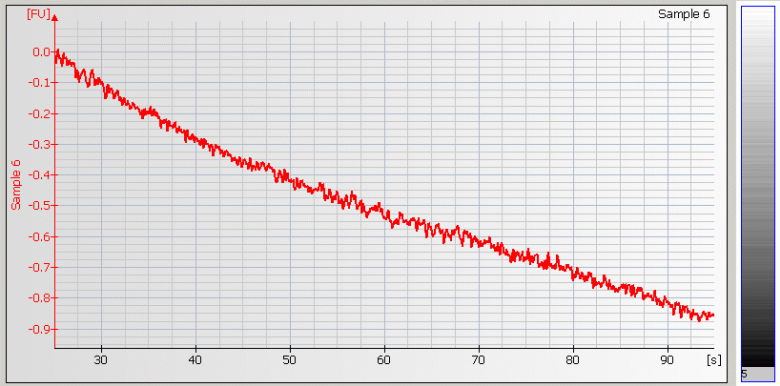
• Wavy Baseline – contamination with genomic DNA |
|
|
|
|
DNA Assays - (back to top)
** General --
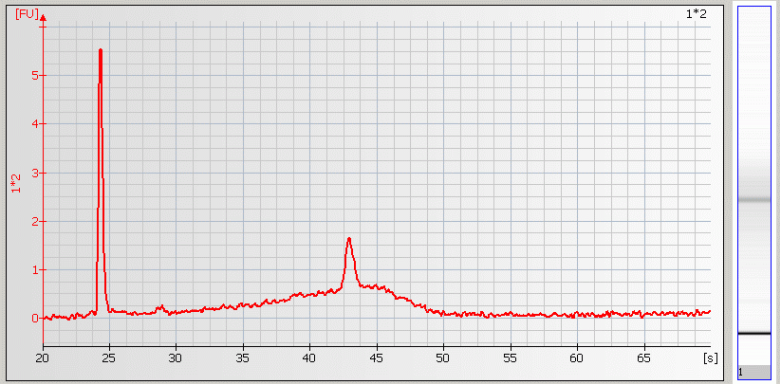
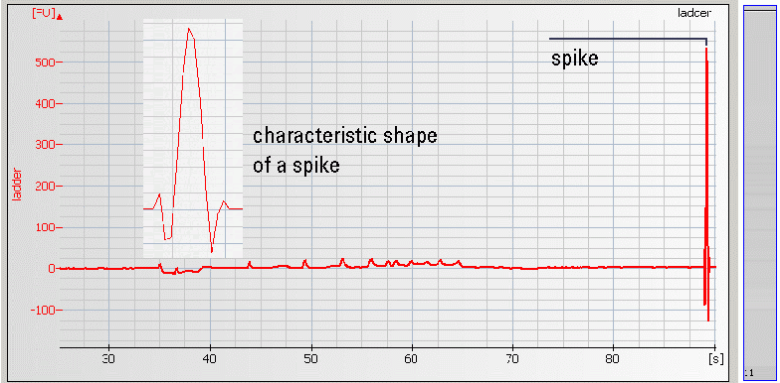
| • Spikes –mechanical spikes are resulted from microparticles or air bubbles present in the system. These spikes, in general, do not affect the quality and/or quantity of the sample. |
|
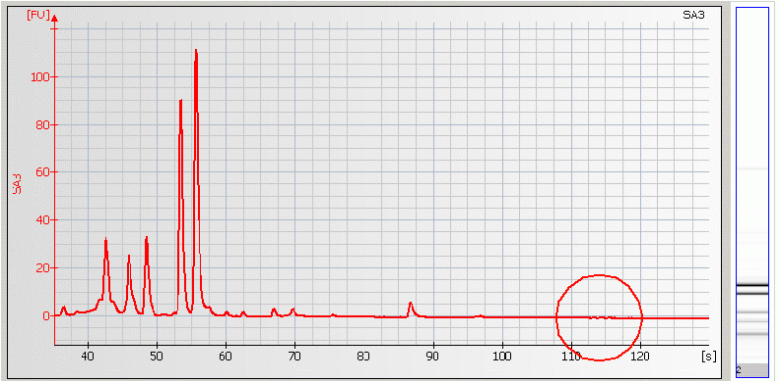
• Missing Upper Marker – may be caused by high salt concentration, ethanol contaminants and/or restriction enzymes present in the sample.
|
|
|
|
| • Missing Peaks – Sample salt concentration is too high |
|
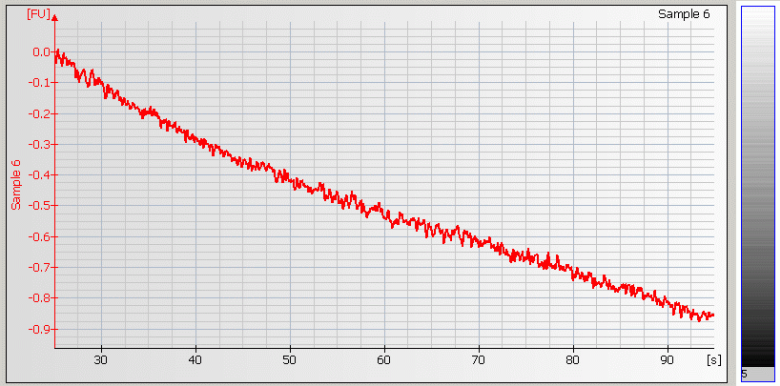
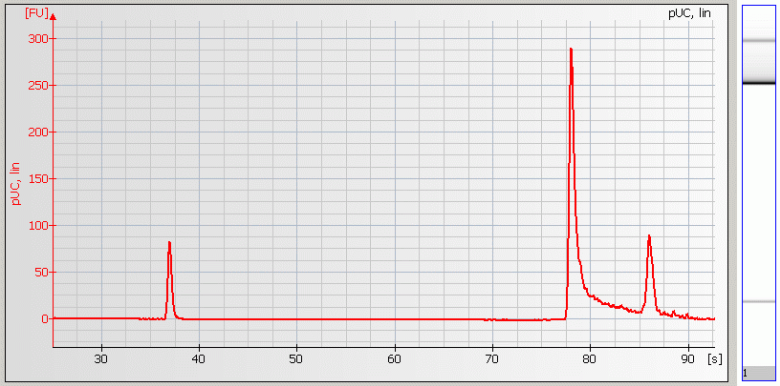
• Baseline Dips – Sample concentration is too high or a contaminant is present |
|
|
|
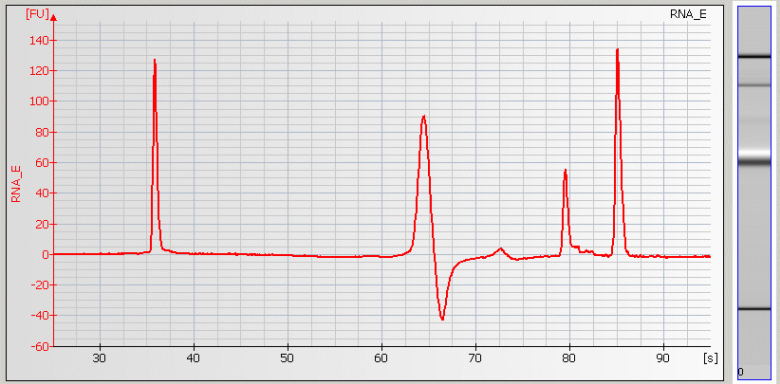
• Peak Tailing – Sample concentration is too high
|
|
|
|
|
|
** NGS Samples -- (back to top)
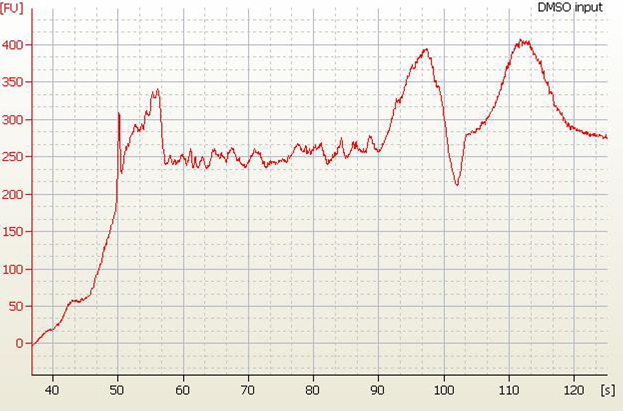
| • High Molecular Weight DNA Contamination |
|
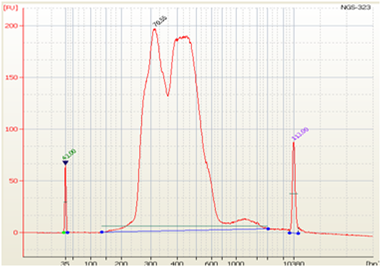
• Split Peaks – sample concentration is too high |
|
|
|
• Excess Adapter - Inefficient ligation due to too much input DNA or the use of incorrect ligation temperature (ligation is performed at 20-25°C)
|
|
• Primer Dimers and PCR Artifacts - |
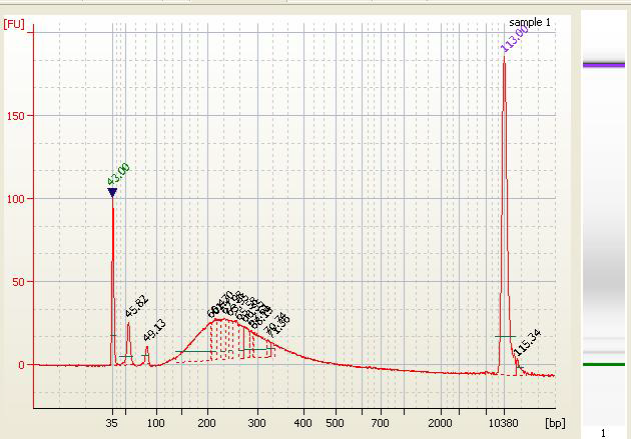
|
|
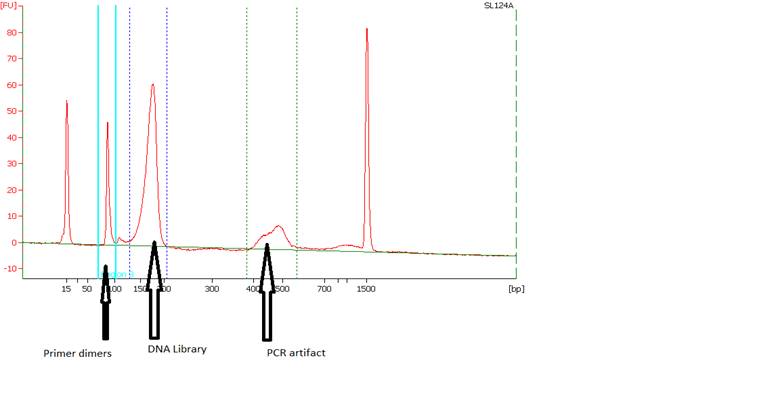
|
• Bead Carried Over –
|
|
• Excess primer - |
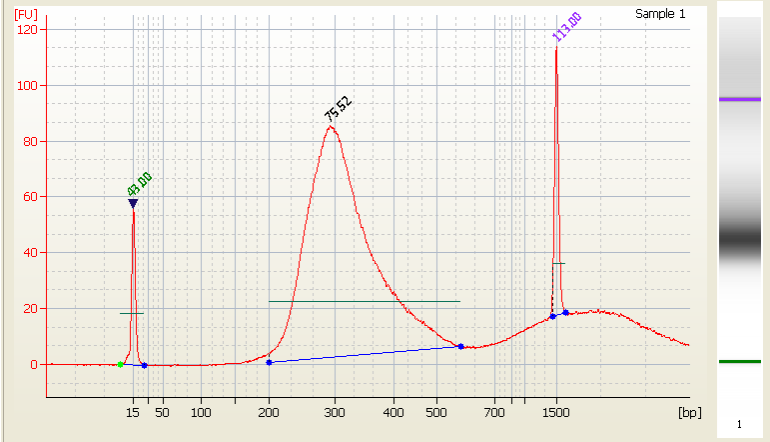
|
|
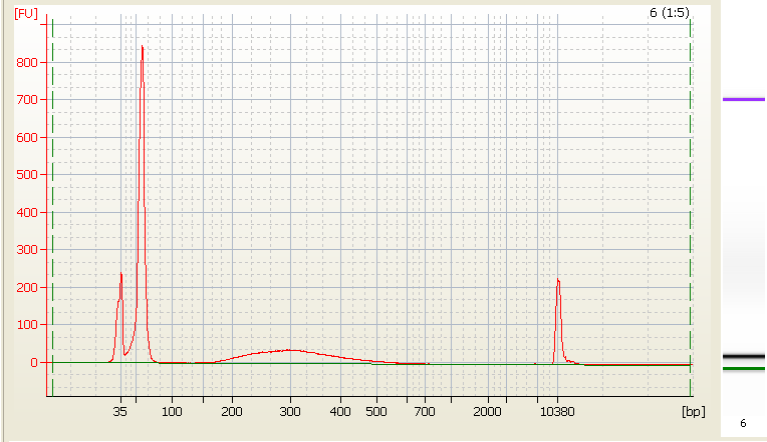
|
|
** References and Helpful Links - The summary above is based on the following resources. You can check them out for more details.
|